Protein cloning, expression and purification
Full-length SV40 LTag (accession number: P03070) N-terminally double 6× histidine-tagged, together with the TEV cleavage site sequence, was cloned into a pFastBac1 plasmid by GenScript. The recombinant expression plasmid was introduced into MultiBac baculoviral DNA in DH10MultiBac, and bacmid DNA was isolated. To prepare the baculovirus, the bacmid DNA of LTag was transfected into Sf9 cells using FuGENE HD (Promega) according to the manufacturer’s instructions. The resulting supernatant was used as the P1 virus stock and amplified to obtain P2 virus stock. The P2 virus stock was then further amplified to obtain P3 virus stock for large-scale expression. LTag was expressed by transfecting 4 l of Sf9 suspension culture at a density of 2 × 106 cells ml−1 with LTag P3 virus for 55–60 h. Cells were collected by centrifugation at 5,500g for 10 min and resuspended in lysis buffer (20 mM Tris (pH 8.0), 50 mM imidazole, 500 mM KCl, 1 mM dithiothreitol (DTT), 5% (v/v) glycerol and EDTA-free protease inhibitor cocktail tablet per 50 ml (Roche, UK)). The cell lysate was clarified by centrifugation at 95,834g for 1 h at 4 °C. The supernatant was directly loaded on to a 5-ml HisTrap affinity column (Cytiva) pre-equilibrated with buffer A (20 mM Tris (pH 8.0), 50 mM imidazole, 500 mM KCl, 1 mM DTT and 10% glycerol). The loaded column was washed with 50 ml of buffer A containing 50 mM imidazole followed by 50 ml of buffer A containing 100 mM imidazole to remove non-specific binding of the protein to the column. Finally, the protein was eluted with a 20-ml gradient to 500 mM imidazole using buffer B containing low salt (20 mM Tris (pH 8.0), 500 mM imidazole, 250 mM KCl, 1 mM DTT, 10 % glycerol). The pooled fractions of LTag were treated with TEV protease to remove the His-tag and dialysed overnight in dialysis buffer (20 mM Tris (pH 8.0), 100 mM KCl, 1 mM DTT and 10% glycerol). The cleaved protein was then loaded into a HisTrap HP 1-ml affinity column (Cytiva) pre-equilibrated with buffer C (20 mM Tris (pH 8), 250 mM KCl, 1 mM DTT and 10% glycerol). The loaded column was then washed with 10 ml of buffer D followed by a 10-ml linear gradient of buffer B. Flow-through fractions that contained LTag were concentrated and loaded on to a HiLoad 16/600 Superdex 200 pg (Cytiva) pre-equilibrated with gel filtration buffer (20 mM Tris (pH 8.0), 100 mM KCl, 1 mM DTT and 10% glycerol). Collected fractions were flash-frozen and stored at −80 °C.
DNA substrates
The forked DNA substrate for reconstitution with LTag and ATP in the absence of Mg2+ was generated by annealing the 5′-GGTGATCGTTCGCTACATGTCGTCAGGATTCCAGGCAG-3′ and 5′-AGACACATGGATGTAGCGAACGATCACC-3′ oligonucleotides, purchased from Eurofins. The forked DNA substrate for reconstitution with LTag and ADP, LTag and ATP in the presence of Mg2+, or LTag and AMP-PNP in the presence of Mg2+ was adapted from SenGupta and Borowiec30, with unpaired arms changed to poly-Ts. The constituent oligonucleotides were therefore 5′-TTCTGTGACTACCTGG ACGACCGGGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′ and 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCCGGTCGTCCAGGTAGTCACAGA-3′. The annealed product was purchased from Eurogentec. The gap DNA substrate for reconstitution of the LTag and ATP apo complex was created by annealing oligonucleotides 5′-AGCTATGACCATGATTACGAATTG[23ddC]-3′ and 5′-TTTTTCGGAGTCGTTTCGACTCCGATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCA ATTCGTAATCATGGTCATAGCT-3′. LTag showed no binding to this substrate without a free 3′ ssDNA tail.
Full SV40 origin DNA (top 5′-CACTACTTCTGGAATAGCTCAGAGGCCGAGGCGGCCTCGGCCTCTGCATAAATAAAAAAAATTA-3′ and bottom 5′-TAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCGGCCTCTGAGCTATTCCAGAAGTAGTG-3′), EP (top 5′-CACTACTTCTGGAATAGCTCAGAGGCCGAGGCG-3′ and bottom 5′-CGCCTCGGCCTCTGAGCTATT CCAGAAGTAGTG-3′) and AT-rich (top 5′-GGCCTCGGCCTCTGCAT AAATAAAAAAAATTA-3′ and bottom 5′-TAATTTTTTTTATTTATGCAGAGGCCGAGGCC-3′) half-origin DNA substrates were synthesized by GenScript (Piscataway) and each annealed by heating at 95 °C for 2 min followed by gentle cooling at −1 °C min−1 to 25 °C. The annealed substrates were further treated with S1 nuclease (Fermentas Life Sciences) at room temperature for 1 h and purified using a PCR clean-up kit (Macherey-Nagel).
Buffers
The buffer used for the LTag–ATP–DNA, LTag–ADP–DNA and LTag–ATP (apo) samples was based on the DNA-binding assay of Li et al.12, comprising 1 mM ATP, 50 mM Tris-HCl (pH 8.0) and 200 mM NaCl. The buffer used for the LTag–ATP-Mg2+–DNA sample was based on the DNA helicase assay from ref. 12 and was composed of 50 mM HEPES (pH 7.5), 1 mM ATP, 3 mM MgCl2, 1 mM DTT and 50 mM NaCl. The buffer used for the LTag–AMP-PNP-Mg2+-DNA complex consisted of 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 100 mM NaCl, 5 mM MgCl2 and 3 mM AMP-PMP. The buffer used for LTag with full- and half-origin DNA contained 50 mM Tris-HCl (pH 8), 100 mM NaCl, 1 mM DTT, 5 mM MgCl2, 5 mM KCl and 3 mM AMP-PNP.
Cryo-EM sample preparation
The LTag stock was preincubated with buffer before addition of DNA substrate (in equimolar ratio to the putative hexamer). The steps were performed in a 4 °C temperature-controlled room. The final protein concentration in samples applied to the grids was approximately 0.098 mg ml−1 (equivalent to 0.2 µM hexamer). R1.2/1.3 or R2/2 UltrAuFoil grids (Quantifoil Micro Tools) were glow-discharged with 40 mA for 5 min inside a Quorum GloQube Plus unit. A graphene oxide (Merck) coating was applied53. Then, 3 µl of sample was applied to the grid in a Vitrobot Mark IV at 4 °C and 100% relative humidity and, following a wait time of 30 s, blotted for 3 s with blot force 4 and plunged into liquid ethane. For the LTag–AMP-PNP-Mg2+–DNA complex, 2 μM LTag (with respect to the hexamer) was mixed in the corresponding buffer and incubated at 4 °C for 30 min. Following this, 2 μM DNA was added, and the reaction was incubated at 4 °C for a further 30 min. Then, 3 µl of sample was deposited on to a R2/2 Au 300 mesh grid previously glow-discharged at 40 mA for 60 s. The grid was frozen on a Vitrobot with a 3-s blotting time, 0-s wait time and a blot force of 5, with 4 °C and 100% humidity in the chamber. To reconstitute the origin melting complexes, LTag stock was mixed with full origin (2:1 molar ratio), EP or AT half origin in equimolar ratio to a final concentration of 1 µM in the aforementioned buffer, and the reaction was incubated at room temperature for 1 h before sample freezing. A Quantifoil Au holey grid (R2/2 300 mesh) was glow-discharged at 30 mA for 30 s using PELCO easiGlow (Ted Pella, Inc.). Then, 3 μl of sample was applied to the glow-discharged grid and blotted for 3 s using a blot force of 0 and no wait time. The sample was then plunge-frozen in liquid ethane using FEI Vitrobot Mark IV preset at 4 °C and 100% humidity.
Cryo-EM data acquisition
LTag–ATP, LTag–ATP–DNA, LTag–ADP–DNA, LTag–AMP-PNP-Mg2+–DNA, and LTag–ATP-Mg2+–DNA data were collected on a Titan Krios G3 transmission electron microscope (Thermo Fisher Scientific) at ×105,000 magnification in AFIS mode with 100-mm objective aperture, using a GATAN K3 direct detection camera at the end of a GATAN BioQuantum energy filter (20 eV slit width) in counted superresolution mode (binning 2), with EPU 2.12 software (Thermo Fisher Scientific), at the Leicester Institute of Structural and Chemical Biology, UK. Individual exposures were acquired at a dose rate of 18 e− px−1 s−1 in 50 equal fractions over 2 s, with a calibrated pixel size of 0.835 Å px−1 and an accumulated dose of 50 e− Å−2. The nominal defocus was varied between −1.0 µm and −2.5 µm in 0.3-µm steps. The gross numbers of micrographs collected from each sample were: 8,072 (LTag–ATP–DNA), 9,737 (LTag–ADP–DNA), 6,127 (LTag–AMP-PNP-Mg2+–DNA) and 8,460 (LTag–ATP-Mg2+–DNA). Data for the LTag–AMP-PNP-Mg2+–DNA, LTag–AMP-PNP-Mg2+–EP, LTag–AMP-PNP-Mg2+–AT and LTag–AMP-PNP-Mg2+–full-origin structures were collected on a Titan Krios G4 transmission electron microscope (Thermo Fisher Scientific) located at the Imaging and Characterization Core Lab of the King Abdullah University of Science and Technology, Saudi Arabia. Micrographs in EER format were recorded at ×130,000 magnification (calibrated pixel size of 0.93 Å px−1) using a Falcon 4i direct detection camera at the end of a Selectris-X energy filter with a 10 eV slit width, with EPU 2.12 software (Thermo Fisher Scientific). Data for LTag–AMP-PNP-Mg2+–DNA were collected with a defocus range of −2.5 to −1.5 μm and an accumulated dose of 38.7 e− Å−2. Data for LTag–AMP-PNP-Mg2+–EP, LTag–AMP-PNP-Mg2+–AT, and LTag–AMP-PNP-Mg2+–full-origin data were collected with a defocus range of −2.6 to −1.1 µm in steps of 0.3 µm, and a total dose of 43 e− Å−2 was applied to the specimen over an exposure time of 5 s.
Cryo-EM data processing
Movie stacks were corrected for beam-induced motion using RELION’s implementation54 of MotionCor2 (ref. 55) with dose-weighting, and contrast transfer function (CTF) estimation of the integrated micrographs was performed using CTFFIND 4.1 (ref. 56). The micrographs were denoised before picking with Topaz v.0.3.0 (ref. 57), both using default models. The picks were extracted using a box size of 384 and subjected to several iterations of two-dimensional classification and selection using VDAM in RELION 4 (ref. 54) (with decreasing binning factors of 4×, 2× and 1×). In general, particle subsets from above were imported into CryoSPARC (Structura Biotechnology) and re-extracted from Patch Motion Correction and Patch CTF Estimation jobs and initially subjected to ab initio reconstruction with three classes (allowing for DNA-bound, apo and junk categories). If two junk classes were detected, the process was repeated using two classes. The resulting particle selections were further corrected for beam-induced motion with local motion correction. Eventual final reconstructions were derived using non-uniform refinement, with CTF and per-particle scale refinements enabled. Refinement masks were constructed with a soft edge (of approximately 20 pixels at half normalized threshold value) around the ordered protein (core helicase) and associated DNA segments of the complex. Gold-standard Fourier shell correlation resolutions were calculated using independently refined random halves of the particle datasets. The LTag–ATP–DNA reconstruction was initially refined to 2.9 Å in RELION 4 using SIDESPLITTER. The final particle subset following two-dimensional classification was subjected to a three-class three-dimensional (VDAM) initial model job. This produced two junk classes; the non-junk class was subjected to two-class three-dimensional classification. This yielded apo (54%) and DNA-bound (44%) classes. The latter was iteratively auto-refined using CTF refinements and Bayesian polishing. A mask was created by thresholding the 15-Å low-pass filtered reconstruction to exclude non-core helicase domains, followed by dilation and padding with 2 and 10 pixels, respectively. The refined particle stack with its assignments was imported into cryoSPARC and rerefined using non-uniform refinement. This gave a better regularized map despite a slightly lower reported global gold-standard Fourier shell correlation resolution. The 3DVA analysis2 of the LTag–ATP-Mg2+–DNA consensus reconstruction was performed using its refinement mask, with three modes and a filter resolution of 5 Å (the global gold-standard Fourier shell correlation resolution being 3 Å), as well as a high-pass resolution of 150 Å. Latent coordinates exhibited a spherical and/or three-dimensional Gaussian distribution, characteristic of continuous conformational variability. Twenty volumes (frame_000 to frame_019) were sampled along each principal component (0, 1, 2). Data had been exhaustively cleaned by repeat classification without separation of conformations. Validation volumes were generated through rolling window partition of particle embeddings centred on the sampled frames along each component, followed by weighted reconstruction using their gold-standard consensus assignments. The 3DVAs of LTag–ADP–DNA and LTag–ATP–DNA were carried out similarly. The LTag–AMP-PNP-Mg2+–DNA structure was wholly refined in RELION 4. In brief, particles picked using Topaz (2,859,203 particles) were extracted with a box size of 256 pixels and 4x downscaling (3.72 Å px−1), and extensively cleaned by two-dimensional classification. The resulting particles (957,358 particles) were subjected to three-dimensional classification with three classes. Particles from the best class (235,707 particles) were re-extracted to full size (0.93 Å px−1) and subjected to three-dimensional refinement, CTF refinement and polishing, resulting in a 3.1-Å resolution map. The refinement mask was created similarly to that used in the LTag–ATP–DNA reconstruction, although it was inclusive of the OBD tier. This was later tightened to the core helicase and DNA for purposes of resolution estimation (and application of global B factor sharpening). LTag–ATP (apo) reconstruction from the gapped DNA sample was refined in RELION. Steps up to and including two-dimensional classification and subset selection were done in RELION 4. Ab initio reconstruction was obtained from CryoSPARC. Three-dimensional classification and auto-refinement were carried out in RELION 5 using Blush regularization (with CTF refinement). Bayesian polishing was not performed. The total numbers of particles incorporated into the final reconstructions were: 92,330 (apo LTag–ATP), 210,910 (LTag–ATP–DNA), 97,587 (LTag–ADP–DNA), 235,707 (LTag–AMP-PNP-Mg2+–DNA) and 201,416 (LTag–ATP-Mg2+–DNA). Local resolution maps were calculated in Phenix and rendered on the full map (before postprocessing) with adjusted colouring. The 3DFSC58 metrics were obtained from the web server at https://3dfsc.salk.edu/.
Origin DNA datasets were processed using CryoSPARC 4.5.3 (Structura Biotechnology). Recorded movies underwent beam-induced motion correction using the Patch Motion Correction module, and CTF estimation was performed with Patch CTF Estimation59. Particles were picked using the Blob Picker with a particle diameter of 130 Å and extracted in a box size of 320 Å. After two rounds of two-dimensional classification, 198,000, 148,881 and 80,452 particles were retained for the EP, AT and full-origin datasets, respectively. These particles were used to generate ab initio reconstructions and refined using CryoSPARC non-uniform refinement. Subsequent CTF refinement and postprocessing resulted in final maps at a resolution of 3.1 Å for half-origin datasets and 3.7 Å for the full-origin dataset. Finally, particle stacks were re-extracted in RELION 5.0-beta-2 and subjected to further three-dimensional classification to identify classes with stronger DNA density.
Model building and refinement
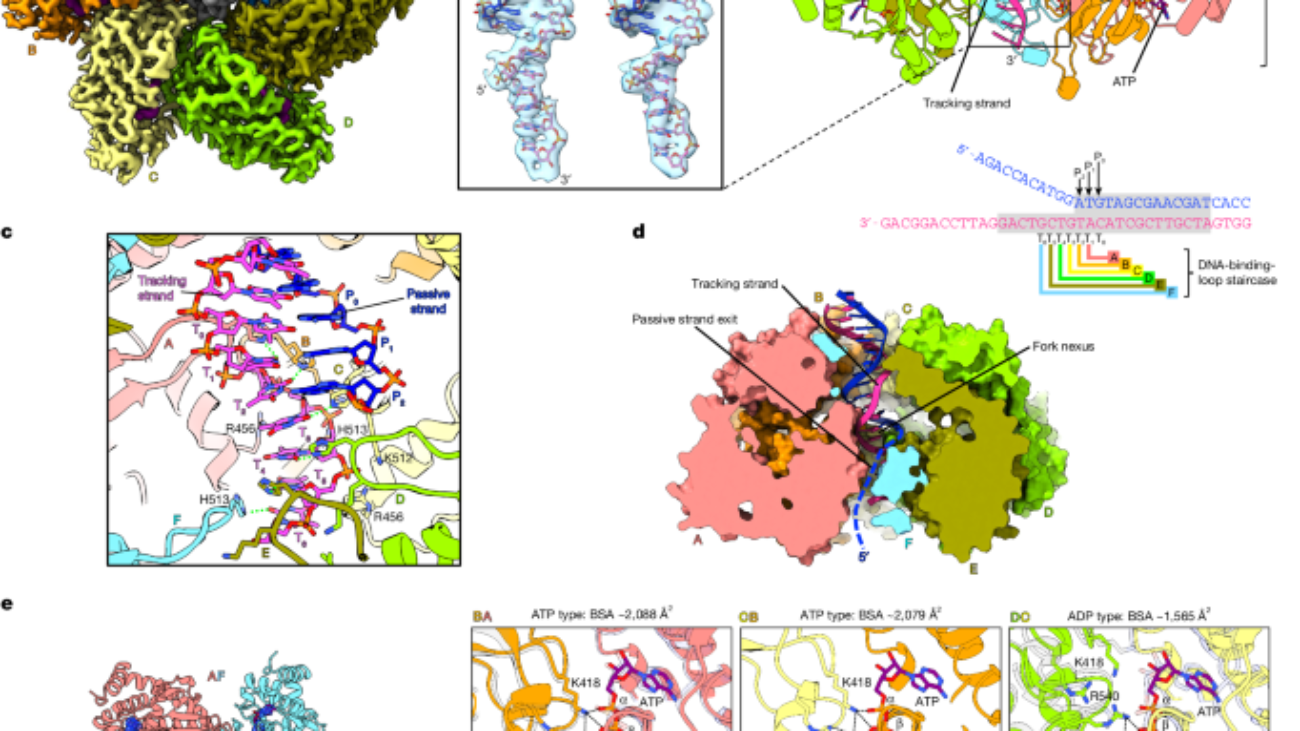
Resulting volumes were postprocessed with EMReady v.1 and v.2 (ref. 28) (using default parameters and without specifying an input structure or mask) for purposes of interactive model-building in ISOLDE29, alongside masked full and/or globally sharpened maps as per automatically calculated B factors. RELION or CryoSPARC postprocessed maps were also used to derive draft models from ModelAngelo (v.0.1)60 to guide the subsequent process. No form of postprocessed maps was used for iterative map refinement itself. For modelling of the LTag–ATP–DNA complex, initially, the crystal structure of ATP-bound LTag (1SVM)11 was real-space refined into the LTag–ATP–DNA map using PHENIX, and oligo-dT6 from the E1–ADP-Mg2+–ssDNA crystal structure (2GXA)25 was fitted into density in ChimeraX. The map–model association was first globally simulated in ISOLDE, followed by multiple cycles of local rebuilding and Phenix real-space refinement with ISOLDE’s recommended settings. In later rounds, further bases in the tracking strand, and complementary strand were added and remodelled. Several iterations of LocScale v.1.0 (ref. 61) and ISOLDE refinement were used to improve the DNA nexus map and model. For modelling of the LTag–ADP–DNA complex, the crystal structure of ADP-bound LTag (1SVL)11 was flexibly fitted into the LTag–ADP–DNA reconstruction using iMODFIT and real-space refined, with ssDNA incorporated as before. The model was then rebuilt and refined as previously. For modelling of the LTag–AMP-PNP-Mg2+–DNA complex, a refined model of LTag–ATP–DNA (omitting the duplex portion of DNA) was initially rigid-body fitted into the LTag–AMP-PNP-Mg2+–DNA reconstruction in ChimeraX. Adaptive distance restraints were applied to individual chains in ISOLDE29 and simulated against the map. Restraints were progressively released and the model fully rebuilt. Towards the end, ATP was replaced with AMP-PNP and Mg2+, resimulated and rebuilt. For modelling of the LTag–ATP-Mg2+–DNA complex into the 3DVA volumes, the LTag–ATP–DNA model, following rigid-body fitting into the LTag–ATP-Mg2+–DNA consensus map, was first rebuilt into a working consensus model. This model was flexibly fitted into the central-most sampled volume (frame_010) from the first principal component. The top-ranked AlphaFold2 (ref. 62) monomer structure prediction for SV40 LTag was superimposed on to each of the six subunits using MatchMaker63. Residues not pertaining to the core helicase 265–627 were deleted. Adaptive distance restraints were applied to individual chains, followed by a whole map–model simulation in ISOLDE. Restraints were progressively released and the model rebuilt end to end. The derived model was placed into two intermediate volumes (frame_005 and frame_015). Adaptive restraints were reapplied and models entirely rebuilt as above. These models were placed into terminal volumes (frame_000 and frame_019), and rebuilt using the same procedure. The central model from principal component 0 was placed into the central volume of component 1. After remodelling, it was used as basis for modelling the two intermediate and then terminal frames as before. The process was repeated for principal component 2. Each residue in all of the above models was manually inspected and/or rebuilt at least once. Q-scores were calculated using the model–map Q-score33 ChimeraX plugin using the default recommended sigma of 0.6. Side-chain-focused model and map validation was carried out using EMRinger v.1.0.0 (ref. 64), averaging a score of 2.9 (well above the requisite 1.0). The consensus LTag–ATP-Mg2+–DNA structure could not be definitively modelled throughout owing to its representing multiple averaged states and was used chiefly as an interim model to generate initial model drafts for variability volumes. Corroborative modelling was performed using Phenix variability refinement65 and assessed using root mean square deviation against the interactively built models, relative to d99 resolution of the maps. For modelling of the LTag–ATP-Mg2+–DNA complex into 3DVA validation volumes, models of terminal frames from each component were rerefined into the back-projected volumes from their corresponding particle subsets in ISOLDE. For modelling of the LTag–ADP–DNA and LTag–ATP–DNA complexes into 3DVA volumes, respective consensus structures were refined into the terminal frames of each component using bulk flexible fitting with adaptive restraints in ISOLDE. For modelling of the LTag–ATP (apo) complex, the LTag–ATP (1SVM) crystal structure was first Phenix real-space refined into the apo LTag–ATP reconstruction postprocessed with EMReady. AlphaFold2 monomer prediction corresponding to amino acids 266–627 was superposed with each of the six chains using MatchMaker and subjected to molecular dynamics bulk flexible fitting with adaptive distance restraints in ISOLDE. Restraints were released inside a whole map–model simulation, followed by localized rebuilding in ISOLDE. For modelling of the LTag–AMP-PNP–full-origin, LTag–AMP-PNP–EP and LTag–AMP-PNP–AT complexes, the LTag–AMP-PNP-Mg2+–DNA fork structure was interactively remodelled into maps of LTag bound to AT and EP half origins using ISOLDE. The paired strand and further tracking-strand bases were added with Coot before refinement in ISOLDE. All final Phenix real-space refined structures were assigned secondary structure annotations using DSSP66. For structures containing AMP-PNP, Mogul ligand restraints downloaded from the Global Phasing Grade Web Server (https://grade.globalphasing.org/) were incorporated into Phenix real-space refinement.
Map and model visualization
Maps were visualized in UCSF Chimera67 and ChimeraX68, and all model illustrations and morphs were prepared using ChimeraX 1.7 and 1.8 or PyMOL 2.6.0.
LTag helicase unwinding assay
EP half-origin primers with 32-nucleotide extensions (5′-CACTACTTCTGGAATAGCTCAGAGGCCGAGGCGGCCTCGGC CTCTGCATAAATAAAAAAAATTA-3′ and 5′-TAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCG GCCTCTGAGCTATTCCA GAAGTAGTG-3′) were first annealed in equivalent portions in an annealing buffer (40 mM Tris-HCl (pH 7.5), 50 mM NaCl and 10 mM MgCl2). The mixture was heated to 95 °C for 5 min then gradually cooled at room temperature at 1 °C per minute. Subsequently, the mixture was subjected to 10% Tris–borate–EDTA polyacrylamide gel electrophoresis at 100 V for 1 h and 15 min. The product was later purified from the gel using ethanol precipitation. The unwinding assay was conducted in a total reaction of 20 μl. Initially, 5 nM of the substrate was incubated with LTag for 10 min in a reaction buffer (20 mM Tris-HCl (pH 7.5), 50 mM KCl, 0.1 mg ml−1 bovine serum albumin, 10 mM MgCl2 and 1 mM DTT) at 37 °C. Various concentrations of LTag were used (50, 100, 200, 300, 400 and 500 nM). The reaction was initiated by addition of 4 mM ATP. After 45 min, the reaction was quenched with stop buffer (1% sodium dodecyl sulfate, 50 mM EDTA and 30% glycerol). Subsequently, 10 μl of the final reaction was loaded on to a 10% Tris–borate–EDTA polyacrylamide gel and run at 100 V for 1 h. Finally, the products were visualized using a Typhoon 9400 laser imager (GE Healthcare).
Statistics and reproducibility
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Source link


Add a Comment